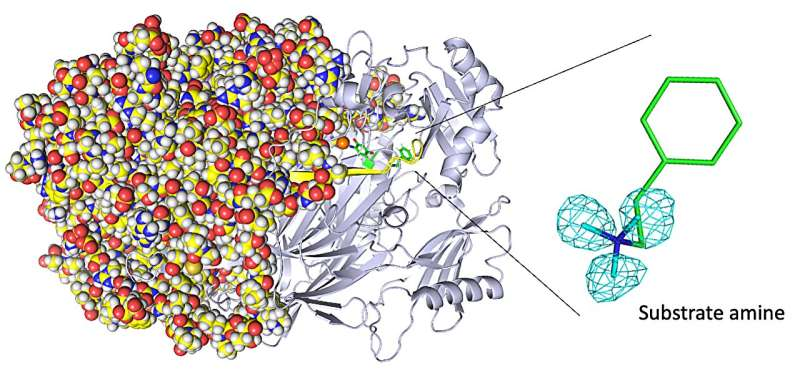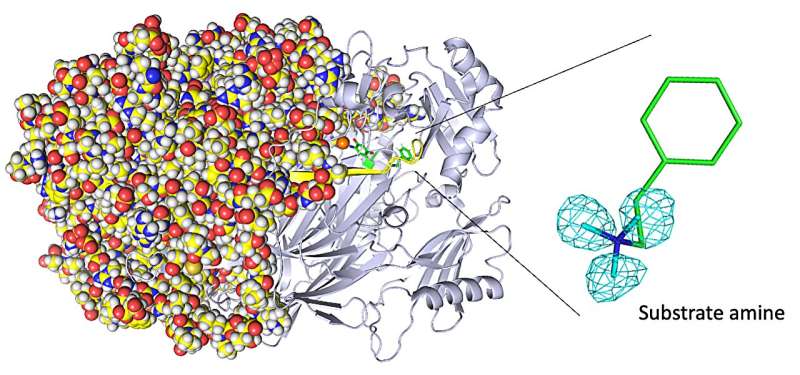
When your wounds heal and your liver detoxifies a poison such as histamine you ingested, you can thank the class of enzymes known as copper amine oxidases for their assistance. Identifying the exact positions of the smallest hydrogen atoms in these enzymes is challenging with commonly used technologies, but is critical to engineering improved enzymes that exhibit unusual yet useful biochemical reactivity.
Now, in a study titled “Neutron crystallography of a semiquinone radical intermediate of copper amine oxidase reveals a substrate-assisted conformational change of the peptidyl quinone cofactor,” published in ACS Catalysis, a team led by researchers at Osaka Medical and Pharmaceutical University and Osaka University has used neutron crystallography to image the atom-by-atom structure of a copper amine oxidase enzyme. This study provides unprecedented structural insights into the enzyme’s biochemistry.
Some copper amine oxidase enzymes exhibit unusual biochemistry, such as quantum tunneling, which enables otherwise inexplicably fast reaction rates. Although it is often challenging to determine the exact position of each hydrogen atom in the enzyme, such knowledge is important for designing corresponding artificial enzymes.
Researchers commonly obtain the atom-by-atom structure of enzymes by X-ray crystallography. However, this technique obtains structural information by diffraction from electrons in the enzyme. Thus, it is insufficient for imaging hydrogen atoms, which generally contain only one electron. Neutron crystallography, which analyzes diffraction from atomic nuclei in the enzyme (all atoms have an atomic nucleus), is an alternative imaging technique that the researchers chose for their work.
“There are pH-dependence, conformational change, and radical intermediate stabilization questions of our enzyme that X-ray crystallography in itself cannot fully explain,” explains Takeshi Murakawa, lead author of the study. “Neutron crystallography is well-suited for answering these questions.”

The researchers obtained numerous insights. For example, they imaged the protonation/deprotonation state (related to the pH) of sites within the enzyme that are important for stabilizing radical species (i.e., especially reactive atoms that contain an unpaired electron). They also characterized the motions of the enzyme’s topaquinone cofactor—sliding, upward tilting, and half-rotation—that facilitate single-electron transfer within the enzyme.
“We disclose binding of a second molecule of high-affinity amine substrate during the enzymatic reaction, a previously unknown event in the enzyme active site,” says Toshihide Okajima, senior author. “X-ray crystallography misses such insights.”
This work has provided previously undisclosed structural details in a copper amine oxidase enzyme that has many functions in biochemical metabolism. Revealing the exact position of the hydrogen atoms in the enzyme helps to explain its efficiency at physiological temperatures and pressures. In the future, researchers might apply these insights to designing artificial enzymes that function used in the chemical industry.
More information: Takeshi Murakawa et al, Neutron Crystallography of a Semiquinone Radical Intermediate of Copper Amine Oxidase Reveals a Substrate-Assisted Conformational Change of the Peptidyl Quinone Cofactor, ACS Catalysis (2023). DOI: 10.1021/acscatal.3c02629
Journal information: ACS Catalysis
Provided by Osaka University