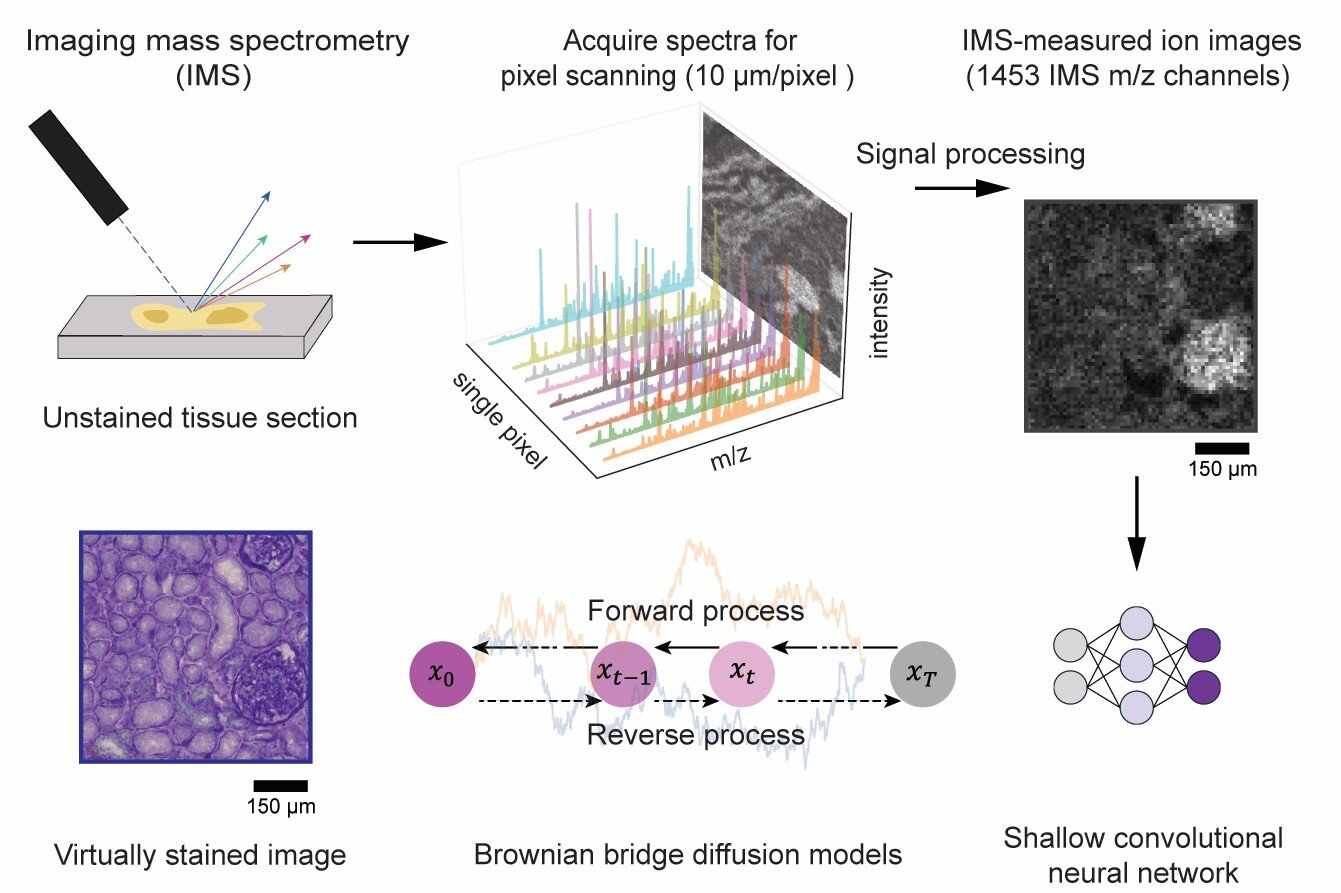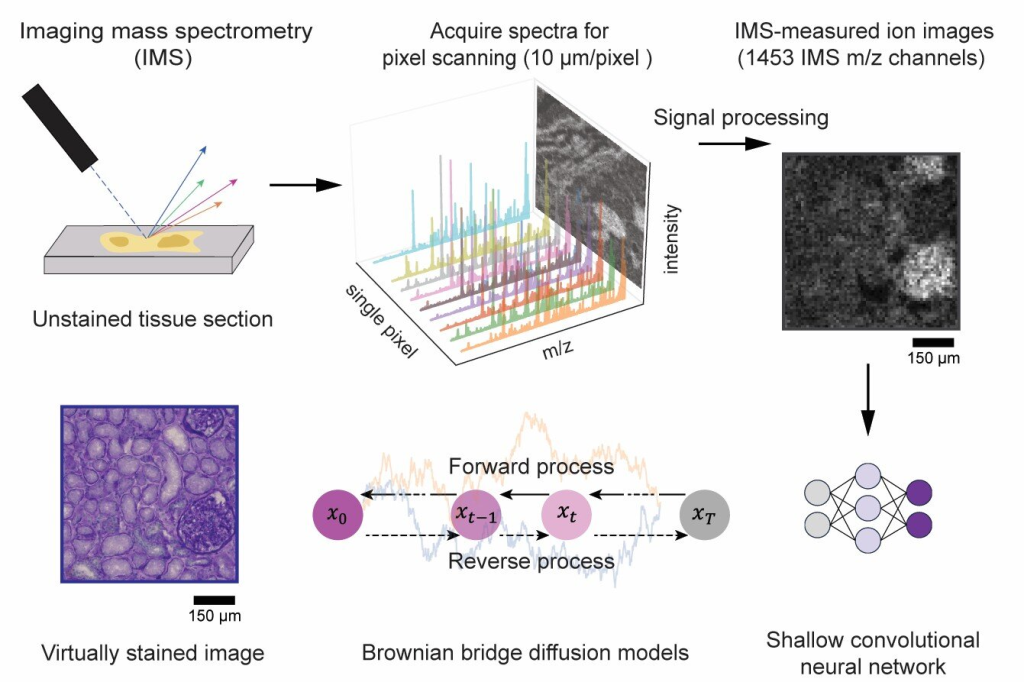
An international team of researchers from the University of California, Los Angeles (UCLA), Vanderbilt University, and Delft University of Technology has developed an artificial intelligence (AI) method that virtually stains images generated through imaging mass spectrometry (IMS). The research is published in the journal Science Advances.
This collaborative effort has achieved significant improvements in spatial resolution and cellular-level detail, all without requiring chemical staining. By leveraging an innovative diffusion-based generative model, the team can digitally produce images comparable to traditional histochemical staining while preserving valuable tissue samples.
Imaging mass spectrometry is a powerful tool capable of mapping hundreds to thousands of molecular species within biological tissues with exceptional chemical specificity. However, conventional IMS is limited by relatively low spatial resolution and a lack of cellular morphological detail, both of which are essential for accurately interpreting molecular profiles within the context of tissue structure.
In this collaborative study, the team introduced a novel diffusion-based virtual staining approach to overcome these challenges. Their method digitally transforms low-resolution, label-free IMS data into high-resolution brightfield microscopy images that closely resemble histochemically stained samples, specifically those stained with Periodic Acid–Schiff (PAS), which highlights polysaccharides, glycoproteins, glycolipids, and mucins in tissues.
Remarkably, the AI framework achieves this despite IMS data having a pixel size nearly 10 times larger than traditional optical microscopy images.
“This diffusion-based approach dramatically enhances the interpretability of mass spectrometry images,” said the corresponding author, Professor Aydogan Ozcan of UCLA. “It virtually introduces microscopic-level histological detail, bridging the gap between molecular specificity and cellular morphology, all without chemically staining the tissue.”
In blind tests on human kidney tissues, the virtually stained images closely matched their chemically stained counterparts, enabling pathologists to accurately identify critical renal structures and disease features directly from the virtual images.
Furthermore, the researchers optimized the noise sampling process during AI inference to ensure highly consistent and reliable staining results, potentially supporting both clinical and research applications.
This technique offers significant benefits for IMS-driven biomedical research and diagnostics, eliminating the need for labor-intensive chemical staining and complex image registration steps. It also preserves tissue integrity for further molecular analyses, thereby streamlining and accelerating mass spectrometry-based molecular histology workflows.
“We envision this approach will open new possibilities in spatial biology and clinical diagnostics,” added Professor Ozcan.
“By digitally generating high-quality histological images from mass spectrometry data alone, we can streamline workflows and potentially advance biomedical discovery.”