Insilico Medicine, a clinical stage generative artificial intelligence (AI)-driven drug discovery company, today announced that it combined two rapidly developing technologies, quantum computing and generative AI, to explore lead candidate discovery in drug development and successfully demonstrated the potential advantages of quantum generative adversarial networks in generative chemistry.
The study, published in the Journal of Chemical Information and Modeling, was led by Insilico’s Taiwan and UAE centers which focus on pioneering and constructing breakthrough methods and engines with rapidly developing technologies—including generative AI and quantum computing—to accelerate drug discovery and development.
The research was supported by University of Toronto Acceleration Consortium director Alán Aspuru-Guzik, Ph.D., and scientists from the Hon Hai (Foxconn) Research Institute.
“This international collaboration was a very fun project,” said Alán Aspuru-Guzik, director of the Acceleration Consortium and professor of computer science and chemistry at the University of Toronto. “It sets the stage for further developments in AI as it meets drug discovery. This is a global collaboration where Foxconn, Insilico, Zapata Computing, and University of Toronto are working together.”
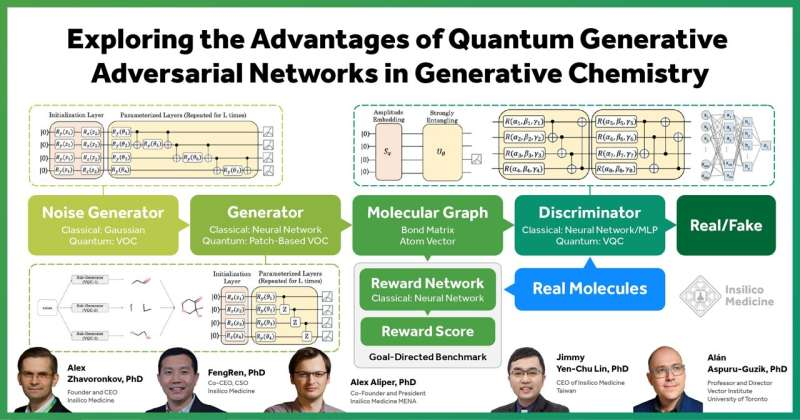
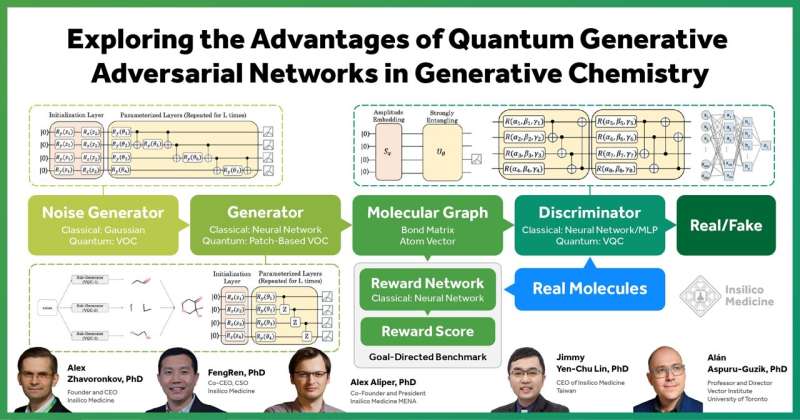
Generative Adversarial Networks (GANs) are one of the most successful generative models in drug discovery and design and have shown remarkable results for generating data that mimics a data distribution in different tasks. The classic GAN model consists of a generator and a discriminator. The generator takes random noises as input and tries to imitate the data distribution, and the discriminator tries to distinguish between the fake and real samples. A GAN is trained until the discriminator cannot distinguish the generated data from the real data.
In this paper, researchers explored the quantum advantage in small molecule drug discovery by substituting each part of MolGAN, an implicit GAN for small molecular graphs, with a variational quantum circuit (VQC), step by step, including as the noise generator, generator with the patch method, and quantum discriminator, comparing its performance with the classical counterpart.
The study not only demonstrated that the trained quantum GANs can generate training-set-like molecules by using the VQC as the noise generator, but that the quantum generator outperforms the classical GAN in the drug properties of generated compounds and the goal-directed benchmark.
In addition, the study showed that the quantum discriminator of GAN with only tens of learnable parameters can generate valid molecules and outperforms the classical counterpart with tens of thousands parameters in terms of generated molecule properties and KL-divergence score.
“Quantum computing is recognized as the next technology breakthrough which will make a great impact, and the pharmaceutical industry is believed to be among the first wave of industries benefiting from the advancement,” said Jimmy Yen-Chu Lin, Ph.D., GM of Insilico Medicine Taiwan and corresponding author of the paper. “This paper demonstrates Insilico’s first footprint in quantum computing with AI in molecular generation, underscoring our vision in the field.”
Building on these findings, Insilico scientists plan to integrate the hybrid quantum GAN model into Chemistry42, the Company’s proprietary small molecule generation engine, to further accelerate and improve its AI-driven drug discovery and development process.
Insilico was one of the first to use GANs in de novo molecular design, and published the first paper in this field in 2016. The Company has delivered 11 preclinical candidates by GAN-based generative AI models and its lead program has been validated in Phase I clinical trials.
“I am proud of the positive results our quantum computing team has achieved through their efforts and innovation,” said Alex Zhavoronkov, Ph.D., founder and CEO of Insilico Medicine. “I believe this is the first small step in our journey. We are currently working on a breakthrough experiment with a real quantum computer for chemistry and look forward to sharing Insilico’s best practices with industry and academia.”
More information: Po-Yu Kao et al, Exploring the Advantages of Quantum Generative Adversarial Networks in Generative Chemistry, Journal of Chemical Information and Modeling (2023). DOI: 10.1021/acs.jcim.3c00562
The data acquisition code and source codes associated with this study are publicly available at: github.com/pykao/QuantumMolGAN-PyTorch
Journal information: Journal of Chemical Information and Modeling
Provided by Insilico Medicine